DeepMind’s AlphaFold joins AI search for new antibiotics

Artificial intelligence (AI) is being used at scale to devise computational models which offer a potentially faster and cheaper way to identify new drugs. But researchers say improvements will have to be made if this approach is to be successful.
Alphabet-owned DeepMind has made headlines in the past with its powerful neural networks which have beaten human players at chess, go and other games of strategy. The company partnered with the European Molecular Biology Laboratory (EMBL) to make a database of predicted protein structure models which was made freely available to the scientific community.
“Our goal at DeepMind has always been to build AI and then use it as a tool to help accelerate the pace of scientific discovery itself, thereby advancing our understanding of the world around us,” says DeepMind Founder and CEO Demis Hassabis.
Now a new study from MIT has explored the potential for existing models to accurately predict interactions between bacterial proteins and antibacterial compounds. Researchers hope this will enable the development of antibiotics, which is essential to work on the antibiotic resistance crisis.
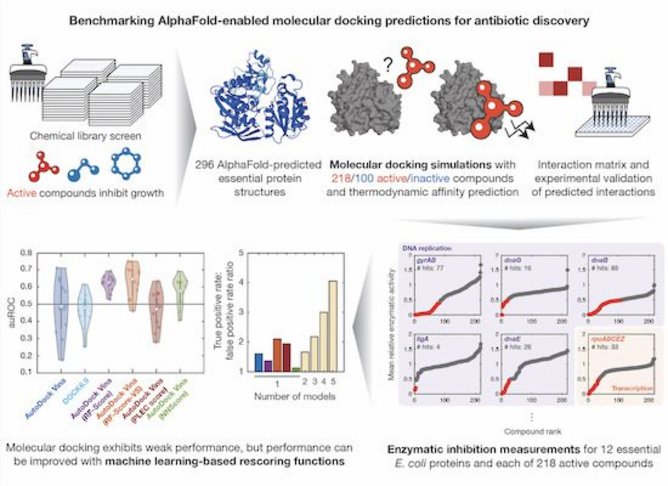
However, researchers found predictions performed little better than chance, with false positive rates similar to true positive rates.
“Breakthroughs such as AlphaFold are expanding the possibilities for in silico drug discovery efforts,” says research leader James Collins, the Termeer Professor of Medical Engineering and Science in MIT’s Institute for Medical Engineering and Science (IMES) and Department of Biological Engineering. “But these developments need to be coupled with additional advances in other aspects of modelling that are part of drug discovery efforts.”
AI and machine learning help in search for antibiotics
Collins and his team studied interactions of 296 proteins with 218 antibacterial compounds, including antibiotics such as tetracyclines. Researchers analysed how compounds interact with proteins using molecular docking simulations, which predict how strongly two molecules will bind together.
Previously this kind of simulation has been successfully used to screen large numbers of compounds against a single protein target. But when researchers attempted to screen many compounds against many potential targets, predictions turned out to be much less accurate.
“The machine-learning models learn not just the shapes, but also chemical and physical properties of the known interactions, and then use that information to reassess the docking predictions,” says MIT postdoc and paper co-author Felix Wong. “We found that if you were to filter the interactions using those additional models, you can get a higher ratio of true positives to false positives.”
Researchers now plan to train the models on more data, including the biophysical and biochemical properties of proteins and their different conformations. Scientists may be able to harness the power of AI-generated protein structures to discover new antibiotics as well as drugs to treat a variety of diseases, including cancer, says Collins.
“We’re optimistic that with improvements to the modelling approaches and expansion of computing power, these techniques will become increasingly important in drug discovery,” says Collins. “However, we have a long way to go to achieve the full potential of in silico drug discovery.”